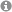Topic
RESEARCH PROJECTS
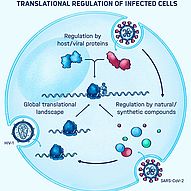
PEERING INTO THE BLACK BOX
Many bacterial and viral pathogens and also their eukaryotic host cells employ non-canonical translation strategies in order to express hidden genes from alternative open reading frames (Caliskan et al., 2015). RNA is a versatile molecule that acts as a key regulator of non-canonical translation events. RNA can exist in various shapes and interact with other regulatory elements such as ncRNAs, small molecules and proteins to alter the meaning of the message encoded in the primary sequence of the mRNA. How RNA structure and regulatory elements drive alternative translation events is currently not fully understood. In addition, it is largely unclear to what extend these translation events are used by the pathogen and the host cell during infections. We use cutting-edge RNA analytics, such as ribosome profiling and deep sequencing combined with single molecule and computational tools to understand dynamics of translation and the functions of RNA regulators during infections. Ultimately, we want to better understand the interplay between the host’s and pathogen’s gene expression and harness our knowledge to develop novel therapeutic strategies to combat infectious diseases.
THE POTATO TOOL: DATA ANALYSIS MADE EASY
The single-molecule technique known as optical tweezers allows probing of intra- and intermolecular interactions that govern complex biological processes. Recent developments have made it easier to collect data, yet it remains difficult to analyze it. To enable high-throughput data analysis, we developed an automated Python-based analysis pipeline called POTATO (practical optical tweezers analysis tool). POTATO uses predefined parameters to automatically process the high-frequency raw data generated by force-ramp experiments and to identify (un)folding events. Our research was published in the Biophysical Journal.
IN FOCUS
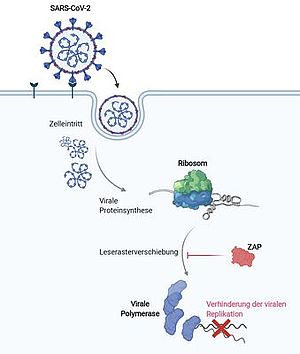
SILVER LININGS IN THE PANDEMIC
Scientists from Neva Caliskan’s research group and other HIRI and HZI labs demonstrate for the first time how ZAP, a protein of the human immune defense system, inhibits the replication mechanism of the SARS-CoV-2 coronavirus and can reduce the viral load by 20-fold. The findings were published in the journal Nature Communications and may help develop antiviral agents in the fight against the pandemic.
Team
Publications
2023
Cis-mediated interactions of the SARS-CoV-2 frameshift RNA alter its conformations and affect function
Pekarek L, Zimmer MM, Gribling-Burrer AS, Buck S, Smyth RP, Caliskan N (2023)
Nucleic Acids Research 51 (2): 728–743
SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9
Schmidt N, Ganskih S, Wei Y, Gabel A, Zielinski S, Keshishian H, Lareau CA, Zimmermann L, Makroczyova J, Pearce C, …, Erhard F, Munschauer M (2023)
Cell 186 (22): 4834-4850.e23
Mouse Liver-Expressed Shiftless Is an Evolutionarily Conserved Antiviral Effector Restricting Human and Murine Hepaciviruses
Zhang Y, Kinast V, Sheldon J, Frericks N, Todt D, Zimmer M, Caliskan N, Brown RJP, Steinmann E, Pietschmann T (2023)
Microbiology Spectrum 11 (4): e0128423
2022
Short- and long-range interactions in the HIV-1 5' UTR regulate genome dimerization and packaging
Ye L, Gribling-Burrer AS, Bohn P, Kibe A, Börtlein C, Ambi UB, Ahmad S, Olguin-Nava M, Smith M, Caliskan N, von Kleist M, Smyth RP (2022)
Nature Structural & Molecular Biology 29 (4): 306-319
Spacer prioritization in CRISPR-Cas9 immunity is enabled by the leader RNA
Liao C, Sharma S, Svensson SL, Kibe A, Weinberg Z, Alkhnbashi OS, Bischler T, Backofen R, Caliskan N, Sharma CM, Beisel CL (2022)
Nature Microbiology 7 (4): 530-541
Editorial: mRNA Translational Control as a Mechanism of Post-transcriptional Gene Regulation
Kiss DL, Vasudevan D, Ho CK, Caliskan N (2022)
Frontiers in Molecular Biosciences 9: 947516
POTATO: Automated pipeline for batch analysis of optical tweezers data
Buck S, Pekarek L, Caliskan N (2022)
Biophysical Journal 121 (15): 2830-2839
Insights from structural studies of the cardiovirus 2A protein
Caliskan N, Hill CH (2022)
Bioscience Reports 42 (1): BSR20210406
Optical Tweezers to Study RNA-Protein Interactions in Translation Regulation
Pekarek L, Buck S, Caliskan N (2022)
Journal of Visualized Experiments (180)
Thinking Outside the Frame: Impacting Genomes Capacity by Programmed Ribosomal Frameshifting
Riegger RJ, Caliskan N (2022)
Frontiers in Molecular Biosciences 9: 842261
2021
Structural and molecular basis for Cardiovirus 2A protein as a viral gene expression switch
Hill CH, Pekarek L, Napthine S, Kibe A, Firth AE, Graham SC, Caliskan N, Brierley I (2021)
Nature Communications 12 (1): 7166
Investigating molecular mechanisms of 2A-stimulated ribosomal pausing and frameshifting in Theilovirus
Hill CH, Cook GM, Napthine S, Kibe A, Brown K, Caliskan N, Firth AE, Graham SC, Brierley I (2021)
Nucleic Acids Research 49 (20): 11938-11958
The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting
Zimmer MM, Kibe A, Rand U, Pekarek L, Ye L, Buck S, Smyth RP, Cicin-Sain L, Caliskan N (2021)
Nature Communications 12 (1): 7193
2020
The SARS-CoV-2 RNA-protein interactome in infected human cells
Schmidt N, Lareau CA, Keshishian H, Ganskih S, Schneider C, Hennig T, Melanson R, Werner S, Wei Y, Zimmer M, …, Bodem J, Munschauer M (2020)
Nature Microbiology 6 (3): 339-353
2019
Thermodynamic control of -1 programmed ribosomal frameshifting
Bock LV, Caliskan N, Korniy N, Peske F, Rodnina MV, Grubmüller H (2019)
Nature Communications 10: 4598
2018
Small synthetic molecule-stabilized RNA pseudoknot as an activator for -1 ribosomal frameshifting
Matsumoto S, Caliskan N, Rodnina MV, Murata A, Nakatani K (2018)
Nucleic Acids Research 46 (16): 8079-8089
2017
Conditional Switch between Frameshifting Regimes upon Translation of dnaX mRNA
Caliskan N, Wohlgemuth I, Korniy N, Pearson M, Peske F, Rodnina MV (2017)
Molecular Cell 66 (4): 558-567.e4
2016
Choreography of molecular movements during ribosome progression along mRNA
Belardinelli R, Sharma H, Caliskan N, Cunha CE, Peske F, Wintermeyer W, Rodnina MV (2016)
Nature Structural & Molecular Biology 23 (4): 342-8
2015
Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting
Caliskan N, Peske F, Rodnina MV (2015)
Trends in Biochemical Sciences 40 (5): 265-74
2014
Programmed -1 frameshifting by kinetic partitioning during impeded translocation
Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV (2014)
Cell 157 (7): 1619-31